Outputs¶
seqcluster¶
counts.tsv: count matrix that can be input of downstream analyses. nloci will be 0 always that the meta-cluster has been resolved successfully. For instance, it can happen that you got sequences you have a bunch of sequences mapping to hundreds of different places on the genome, then seqcluster doesn’t resolve that, and put everything under the larger region covered by those sequences. So, mainly, 0 all are good rows. The ann column is just where the meta-clusters overlap with. It can happen that one name appears many times if different locations of the meta-cluster map to different copies of that feature. OR if the annotation file used had multiple lines for that.read_stats.tsv: number of reads for each sample after each step in the analysis. Meant to give a hint if we lose a lot of information or not.size_counts.tsv: size distribution of the small RNA by annotation group. (position, reads, cluster)seqcluster.json: json file containing all information. This file is used as the input of the report suit.log/run.log: all messages at debug levellog/trace.log: to keep trace of algorithm decisions
Report¶
Beside the static HTML report that you can get using report subcommand, you can download this HTML. (watch the repository to get notifications of new releases.)
- Go inside
seqclusterVizfolder - Open
reader.html - Upload the
seqcluster.dbfile generated byreportsubcommand. - Start browsing your data!
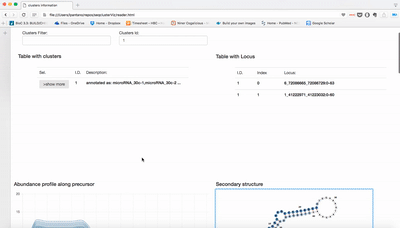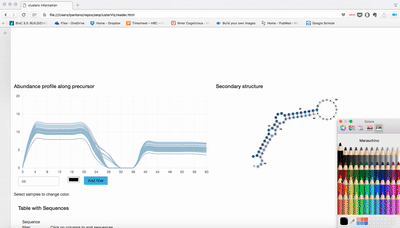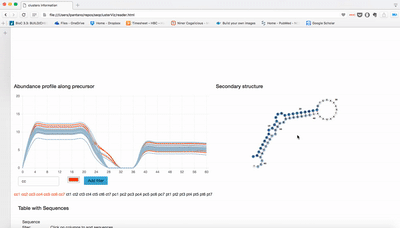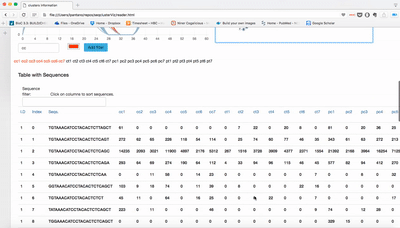
Meaning of different sections:
- Top-left table shows list of meta-clusters, user can filter by number ID or keywords.
- Top-right table shows positions where this meta-cluster has been detected.
- Expression profile along precursor: Lines are number of reads in that position of the precursor. It is sum of the log2 RPM of the expression for each sample.
- Table: raw counts for each sample and sequence. Only top 100 are shown.
- secondary structure: The region with more sequences inside meta-cluster is used to plot the secondary structure. Colors refers to abundance in each position. Darker means more abundance.
An example of the HTML code: